Help page |
|
|
| Annotation |
|
| SPM |
| CTM |
| DPM |
| RPM |
| Similarity Measure |
Correlation Coefficient |
| Specific Genes |
| Selective Genes |
| Housekeeping Genes |
| Repressed Genes |
| |
| User Guide |
| |
| Upload |
| Expression Cutoff |
| Gene Search |
| Pattern Gene Search |
| Similarity/Correlation Analysis |
| Download |
|
|
SPM
SPM is a statistical parameter introduced as a sensitive indicator in quantitative estimation of gene expression patterns.
Each gene expression profile of gene x is transformed into a vector Xp:
|
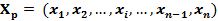
|
where n is the number of samples in the profile and xi is the gene expression level in sample i. For each element in the profile Xp, the expression xi can also be represented by a vector Xi in high dimension feature space:
|
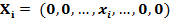
|
The SPM of a gene in a sample was then determined by calculating the cosine value of intersection angle Θ between vector Xi and Xp in high dimension feature space:
|
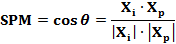
|
Theoretically, the SPM ranges from 0 to 1.0. A value close to 1.0 indicates that element xi is the major contributor to the length of profile Xp in high dimension feature space; in biological term, high sample specificity.
|
CTM
The Contribution Measure (CTM) is a parameter developed to help identification of selective genes. The CTM was calculated by: |
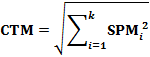
|
Theoretically, the CTM ranges from 0 to 1.0. In biology, a CTM value close to 1.0 suggests gene expressions are enriched in selective samples (normally 2 ≤ SampleNum ≤ 4) comparing to that in other samples. |
DPM
The DPM, in principle, is to map the standard deviation of a gene expression profile into a region of 0 to 1.0. The gene expression profile (Xp) is first converted to its corresponding SPM profile (XSPM): |
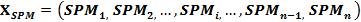
|
Then, the DPM is determined by: |
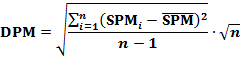
|
Theoretically, the DPM ranges from 0 to 1.0. In biology, a DPM value close to 0 suggests that gene evenly expresses in all samples of a gene expression profile. |
RPM
The Repression Measure (RPM) is a parameter for identification of repressed genes.The gene expression profile in multiple samples were roughly divided into two groups according to their expression levels: the putative repressed expressions and the expressions. Then the RPM was calculated by: |
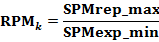
|
where the "SPMrep_max" stands for the maximum SPM value in the putative repressed expression group and "SPMexp_min" stands for minimun SPM value in the expression group, and k is the number of repressed samples.
Theoretically, the RPM ranges from 0 to 1.0. A RPM value close to 0 indicates high degree of difference between repression group and expression group. |
Similarity Measure |
Gene X: |
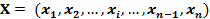 |
Gene Y: |
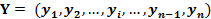 |
Similarity Measure: |
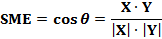 |
Theoretically, the SME ranges from 0 to 1.0. In biology, SME indicates how similar two gene distribution curves are. A SME value close to 1 means the high similarity of two distributions. |
Correlation Coefficient |
| Correlation Coefficient: |
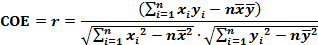 |
Theoretically, the COE (r) ranges from -1 to 1. A value of correlated coefficient r close to 1 or -1 concludes the high correlation of two distributions statistically, while co-expression (close to 1) or inverse-expression (close to -1) in biological extent. |
Specific Genes
Genes are specifically expressed in only one sample. Noramlly, specific gene has a high SPM value, e.g. SPM ≥ 0.9. |
Selective Genes
The selective genes are a group of genes whose expressions are enriched in only several conditions (samples). Normally, gene expressions are enriched in limited (e.g. 2 ≤ SampleNum ≤ 4) samples (e.g. CTM ≥ 0.9 and DPM ≥ 0.9), and expressions in each of the selective samples are comparatively high (e.g. SPMi ≥ 0.4). |
Housekeeping Genes
Housekeeping genes are generally defined as genes that express ubiquitously in all conditions. In strict sense, housekeeping genes evenly express in all samples (e.g. DPM ≤ 0.3). |
Repressed Genes
Genes are expressed in almost all conditions except in one or several conditions. |
|
| |
| |
|